Using String Distance {stringdist} To Handle Large Text Factors, Cluster Them Into Supersets
Practical walkthroughs on machine learning, data exploration and finding insight.
Resources
Packages Used in this Walkthrough
- {stringdist} - feature selection for ensembles
- {RCurl} - machine learning tools
If you’re wondering whether you’re getting the most out of a text-based, factor variable from a large data set, then you’re not alone. There are so many ways of deconstructing text variables. If every entry is made of repeated text from a small set of possibilities, then dummifying it is the easiest way to proceed. On the other hand, if every entry is unique, then resorting to Natural Language Processing (NLP) may be required. This article tackles the gray area in between, where the data is neither unique nor small, where dummifying won’t work yet NLP may still be avoided.
So that we are on the same page, imagine a data set with 10 million rows with at least one feature/column being a text-based factor. It is not made up of free-text, where every entry is unique, instead, it is made up of repeated text: for example 10,000 possibilities repeated over 10 million rows. This would be hard to dummify, as it will blow up your feature space, and would take forever to group by hand.
What Is One To Do?
- We could encode the text entries as integers or binaries and hope for the best (as it is not ordinal in nature, linear models would suffer but classification models would be OK).
- We could take the top X most popular ones and overwrite the rest as 'other' and dummify the resulting set (I have used that method many times and will write up a post on the subject).
- But a more interesting approach that affords much less loss of information, is grouping them into supersets and the subject of this walkthrough.
Grouping With {stringdist}
Could those 10,000 possibilities mentioned earlier be grouped into a superset representing only a tenth or a fifth of its original size? What is close to impossible to do by hand is trivial with string distance:
...a metric that measures distance ("inverse similarity") between two text strings for approximate string matching or comparison and in fuzzy string searching. (Source: Wikipedia)
The {strndist} package offers ‘Approximate string matching and string distance functions’. It offers many algorithms but the two I found the most interesting for short sets of words are:
...the Jaro–Winkler distance (Winkler, 1990) is a measure of similarity between two strings. The higher the Jaro–Winkler distance for two strings is, the more similar the strings are. The Jaro–Winkler distance metric is designed and best suited for short strings such as person names. The score is normalized such that 0 equates to no similarity and 1 is an exact match. (Source: Wikipedia)
and
...the Levenshtein distance between two words is the minimum number of single-character edits (i.e. insertions, deletions or substitutions) required to change one word into the other. (Source: Wikipedia)
Hi there, this is Manuel Amunategui- if you're enjoying the content, find more at ViralML.com
Let’s Code!
Enough chitchat, let’s download the vehicles data set from Hadley Wickham hosted on Github. It is a big and diverse data set, perfect for our needs:
library(RCurl)
urlfile <-'https://raw.githubusercontent.com/hadley/fueleconomy/master/data-raw/vehicles.csv'
x <- getURL(urlfile, ssl.verifypeer = FALSE)
vehicles <- read.csv(textConnection(x))
# alternative way of getting the data if the above snippet doesn't work:
# urlData <- getURL('https://raw.githubusercontent.com/hadley/fueleconomy/master/data-raw/vehicles.csv')
# vehicles <- read.csv(text = urlData)
We’re going to focus on one single feature in the data set: model. Let’s start with some basic statistics on that feature to understand what we’re dealing with:
nrow(vehicles)
[1] 34631
length(unique(vehicles$model))
[1] 3234
So, we have a data set of over 34,631 vehicles, but model is comprised of only 3,234 unique model names repeated through out all the observations/rows. Let’s look at the first 100 rows so we don’t get overwhelmed with too much the data:
vehicles_small <- vehicles[1:100,]
Out of those 100 observations, model has only 45 unique model names and here is a small sample of what it holds:
length(unique(vehicles_small$model))
[1] 45
head(unique(as.character(vehicles_small$model)))
[1] "Spider Veloce 2000" "Testarossa" "Charger"
[4] "B150/B250 Wagon 2WD" "Legacy AWD Turbo" "Loyale"
Let’s run some basic string distance on this subset by calling the stringdistmatrix function to see how it can help us tame the model variable into smaller supersets. In its simplest form, the function stringdistmatrix only requires a unique set of text values and the method to cluster the data:
stringdistmatrix(a, b, method = c("osa", "lv", "dl", "hamming", "lcs",
"qgram", "cosine", "jaccard", "jw", useBytes = FALSE,
weight = c(d = 1, i = 1, s = 1, t = 1), maxDist = Inf, q = 1, p = 0,
useNames = FALSE, ncores = 1, cluster = NULL)
We’ll pass it the unique list of models, request the Jaro–Winkler distance algorithm (my favorite for this task), cluster the results into 20 groups with the hclust function and plot the resulting dendrogram:
library(stringdist)
uniquemodels <- unique(as.character(vehicles_small$model))
distancemodels <- stringdistmatrix(uniquemodels,uniquemodels,method = "jw")
rownames(distancemodels) <- uniquemodels
hc <- hclust(as.dist(distancemodels))
plot(hc)
rect.hclust(hc,k=20)
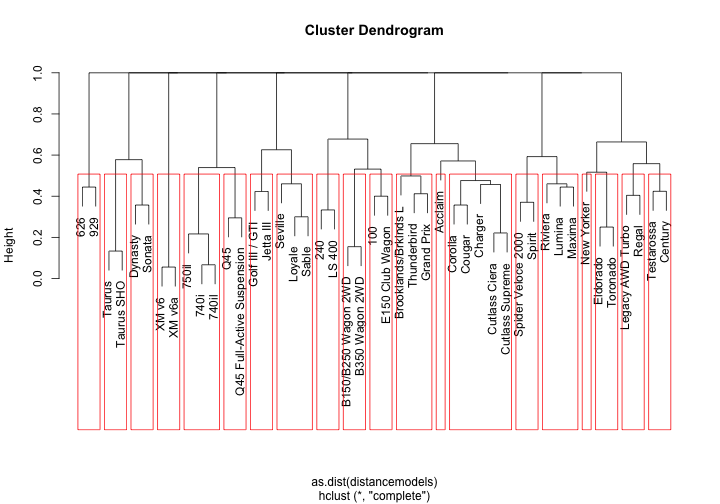
stringdistmatrix works in tandem with hclust, one creates the model, the other enforces the clusters. Our algorithm created a Wagon and Taurus group along with some number groups. So far it isn’t extremely impressive as its only using a small subset of data - wait till we open things up!
We now look at a bigger subset of the vehicles data set. Let’s pull the first 2000 observations:
vehicles_small <- vehicles[1:2000,]
length(unique(vehicles_small$model))
[1] 481
Out of those 2000 observations, we have 481 unique model names. Let’s ask stringdistmatrix to group those into 200 groups:
uniquemodels <- unique(as.character(vehicles_small$model))
distancemodels <- stringdistmatrix(uniquemodels,uniquemodels,method = "jw")
rownames(distancemodels) <- uniquemodels
hc <- hclust(as.dist(distancemodels))
dfClust <- data.frame(uniquemodels, cutree(hc, k=200))
names(dfClust) <- c('modelname','cluster')
Let’s visualize the quantities of models in each group created by the Jaro–Winkler distance algorithm:
plot(table(dfClust$cluster))
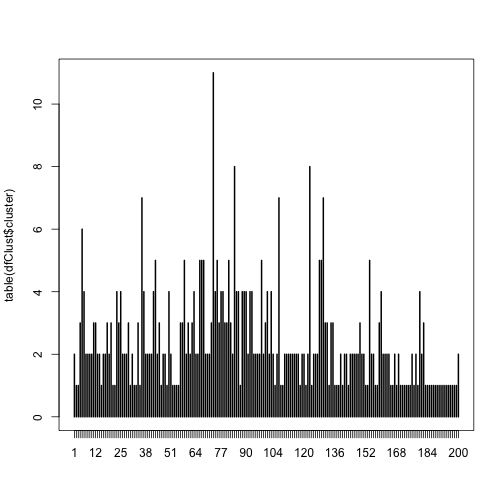
print(paste('Average number of models per cluster:', mean(table(dfClust$cluster))))
## [1] "Average number of models per cluster: 2.405"
The largest cluster contains over 10 models but the average is 2.4 models per cluster. Now, lets look at the top groups and see what the algorithm did (don’t sweat this code, it simply orders the data by cluster size):
t <- table(dfClust$cluster)
t <- cbind(t,t/length(dfClust$cluster))
t <- t[order(t[,2], decreasing=TRUE),]
p <- data.frame(factorName=rownames(t), binCount=t[,1], percentFound=t[,2])
dfClust <- merge(x=dfClust, y=p, by.x = 'cluster', by.y='factorName', all.x=T)
dfClust <- dfClust[rev(order(dfClust$binCount)),]
names(dfClust) <- c('cluster','modelname')
head (dfClust[c('cluster','modelname')],50)
## cluster modelname
## 192 73 K1500 Pickup 4WD
## 191 73 S10 Pickup 2WD
## 190 73 W250 Pickup 4WD
## 189 73 F150 Pickup 2WD
## 188 73 S10 Pickup 4WD
## 187 73 D100/D150 Pickup 2WD
## 186 73 F250 Pickup 2WD
## 185 73 C1500 Pickup 2WD
## 184 73 F150 Pickup 4WD
## 183 73 D250 Pickup 2WD
## 182 73 W100/W150 Pickup 4WD
## 341 123 Postal Cab Chassis 2WD
## 340 123 S10 Cab Chassis 2WD
## 339 123 Dakota Cab Chassis 2WD
## 338 123 Cab/Chassis 2WD
## 337 123 Cab Chassis 2WD
## 336 123 S15 Cab Chassis 2WD
## 335 123 Truck Cab Chassis 2WD
## 334 123 D250 Cab Chassis 2WD
## 236 84 Yukon 1500 4WD
## 235 84 Suburban C10 2WD
## 234 84 SJ 410V 4WD
## 233 84 Suburban 1500 2WD
## 232 84 Suburban K10 4WD
## 231 84 Yukon K1500 4WD
## 230 84 SJ 410 4WD
## 229 84 Suburban 1500 4WD
## 365 130 900 Convertible
## 364 130 318i Convertible
## 363 130 Convertible
## 362 130 XJS V12 Convertible
## 361 130 E320 Convertible
## 360 130 325i Convertible
## 359 130 XJS Convertible
## 307 107 Sidekick 2Door 2WD
## 306 107 Sidekick Hardtop 2WD
## 305 107 Sidekick 4Door 2WD
## 304 107 Sidekick 2Door 4WD
## 303 107 Sidekick 2WD
## 302 107 Sidekick Hardtop 4WD
## 301 107 Sidekick 4Door 4WD
## 86 36 240 Wagon
## 85 36 E320 Wagon
## 84 36 940 Wagon
## 83 36 850 Wagon
## 82 36 960 Wagon
## 81 36 E150 Club Wagon
## 80 36 100 Wagon
## 13 5 Legacy AWD Turbo
## 12 5 Legacy Wagon
Out of the 200 clusters we requested, cluster 73 is the largest holding 11 models. Clearly, it picked up on the word pickup flanked by two words on either side with the right one being 2WD or 4WD. Cluster 123 looked for Cab Chassis, even picking up a Cab/Chassis in the process. You get the idea and, hopefully, are impressed with how a few lines of code reduced 2000 observations into 200 groups. The exact same process would apply to 20,000 observations or 20 million…
Creating New Variables Through Combining Multiple Features
An offshoot of this process is to create new groups by combining existing features and running the results through stringdistmatrix. Let’s try combining model with trany:
vehicles_small$modelAndTrany <- paste0(as.character(vehicles_small$model)," ",as.character(vehicles_small$trany))
print(length(unique(vehicles_small$modelAndTrany)))
## [1] 808
Our new field has 808 unique values out of our 2000 small_vehicles data frame. Let’s run it through the Jaro–Winkler distance algorithm, request 500 clusters and check out the top groups:
uniquemodels <- unique(as.character(vehicles_small$modelAndTrany))
distancemodels <- stringdistmatrix(uniquemodels,uniquemodels,method = "jw")
rownames(distancemodels) <- uniquemodels
hc <- hclust(as.dist(distancemodels))
dfClust <- data.frame(uniquemodels, cutree(hc, k=500))
names(dfClust) <- c('modelname','cluster')
t <- table(dfClust$cluster)
t <- cbind(t,t/length(dfClust$cluster))
t <- t[order(t[,2], decreasing=TRUE),]
p <- data.frame(factorName=rownames(t), binCount=t[,1], percentFound=t[,2])
dfClust <- merge(x=dfClust, y=p, by.x = 'cluster', by.y='factorName', all.x=T)
dfClust <- dfClust[rev(order(dfClust$binCount)),]
names(dfClust) <- c('cluster','modelname')
head (dfClust[c('cluster','modelname')],50)
## cluster modelname
## 38 16 960 Automatic 4-spd
## 37 16 90 Automatic 4-spd
## 36 16 940 Automatic 4-spd
## 35 16 900 Automatic 4-spd
## 34 16 E500 Automatic 4-spd
## 33 16 100 Automatic 4-spd
## 32 16 9000 Automatic 4-spd
## 31 16 850 Automatic 4-spd
## 27 14 G20 Automatic 4-spd
## 26 14 240 Automatic 4-spd
## 25 14 S420 Automatic 4-spd
## 24 14 240SX Automatic 4-spd
## 23 14 C280 Automatic 4-spd
## 22 14 C220 Automatic 4-spd
## 21 14 E420 Automatic 4-spd
## 221 120 960 Wagon Automatic 4-spd
## 220 120 E320 Wagon Automatic 4-spd
## 219 120 850 Wagon Automatic 4-spd
## 218 120 240 Wagon Automatic 4-spd
## 217 120 100 Wagon Automatic 4-spd
## 216 120 940 Wagon Automatic 4-spd
## 185 99 SW Manual 5-spd
## 184 99 S4 Manual 5-spd
## 183 99 S6 Manual 5-spd
## 182 99 NSX Manual 5-spd
## 181 99 SL Manual 5-spd
## 180 99 SC Manual 5-spd
## 248 129 Ram 1500 Pickup 4WD Manual 5-spd
## 247 129 Ram 2500 Pickup 2WD Manual 5-spd
## 246 129 Ram 2500 Pickup 4WD Manual 5-spd
## 245 129 Ram 1500 Pickup 2WD Manual 5-spd
## 244 129 Ram 50 Pickup 2WD Manual 5-spd
## 243 128 Ram 1500 Pickup 2WD Automatic 4-spd
## 242 128 Ram 2500 Pickup 4WD Automatic 4-spd
## 241 128 Ram 50 Pickup 2WD Automatic 4-spd
## 240 128 Ram 1500 Pickup 4WD Automatic 4-spd
## 239 128 Ram 2500 Pickup 2WD Automatic 4-spd
## 177 97 NSX Automatic 4-spd
## 176 97 SC Automatic 4-spd
## 175 97 SVX Automatic 4-spd
## 174 97 SW Automatic 4-spd
## 173 97 SL Automatic 4-spd
## 154 83 SL600 Automatic 4-spd
## 153 83 500SEL Automatic 4-spd
## 152 83 SL500 Automatic 4-spd
## 151 83 400SEL Automatic 4-spd
## 150 83 500SL Automatic 4-spd
## 47 18 540i Automatic 5-spd
## 46 18 840ci Automatic 5-spd
## 45 18 740il Automatic 5-spd
Conclusion
stringdistmatrix is a very flexible function with many tunable features. The cluster size, the algorithm, the concatenation of text with text and/or numbers create numerous and mind-boggling possibilities. Even with all these settings it is still so much easier than creating supersets by hand! String distance matching is case sensitive so you may get better groups if you force all text to once case. Have fun with this…
Full source code (also on GitHub):
# get the Hadley Wickham's vehicles data set
library(RCurl)
urlfile <-'https://raw.githubusercontent.com/hadley/fueleconomy/master/data-raw/vehicles.csv'
x <- getURL(urlfile, ssl.verifypeer = FALSE)
vehicles <- read.csv(textConnection(x))
# size the data
nrow(vehicles)
length(unique(vehicles$model))
# get a small sample for starters
vehicles_small <- vehicles[1:100,]
length(unique(vehicles_small$model))
head(unique(as.character(vehicles_small$model)))
# call the stringdistmatrix function and request 20 groups
library(stringdist)
uniquemodels <- unique(as.character(vehicles_small$model))
distancemodels <- stringdistmatrix(uniquemodels,uniquemodels,method = "jw")
rownames(distancemodels) <- uniquemodels
hc <- hclust(as.dist(distancemodels))
# visualize the dendrogram
plot(hc)
rect.hclust(hc,k=20)
# get a bigger sample
vehicles_small <- vehicles[1:2000,]
length(unique(vehicles_small$model))
# run the stringdistmatrix function and request 200 groups
uniquemodels <- unique(as.character(vehicles_small$model))
distancemodels <- stringdistmatrix(uniquemodels,uniquemodels,method = "jw")
rownames(distancemodels) <- uniquemodels
hc <- hclust(as.dist(distancemodels))
dfClust <- data.frame(uniquemodels, cutree(hc, k=200))
names(dfClust) <- c('modelname','cluster')
# visualize the groupings
plot(table(dfClust$cluster))
print(paste('Average number of models per cluster:', mean(table(dfClust$cluster))))
# lets look at the top groups and see what the algorithm did:
t <- table(dfClust$cluster)
t <- cbind(t,t/length(dfClust$cluster))
t <- t[order(t[,2], decreasing=TRUE),]
p <- data.frame(factorName=rownames(t), binCount=t[,1], percentFound=t[,2])
dfClust <- merge(x=dfClust, y=p, by.x = 'cluster', by.y='factorName', all.x=T)
dfClust <- dfClust[rev(order(dfClust$binCount)),]
names(dfClust) <- c('cluster','modelname')
head (dfClust[c('cluster','modelname')],50)
# try combining fields together
vehicles_small$modelAndTrany <- paste0(as.character(vehicles_small$model)," ",as.character(vehicles_small$trany))
print(length(unique(vehicles_small$modelAndTrany)))
uniquemodels <- unique(as.character(vehicles_small$modelAndTrany))
distancemodels <- stringdistmatrix(uniquemodels,uniquemodels,method = "jw")
rownames(distancemodels) <- uniquemodels
hc <- hclust(as.dist(distancemodels))
dfClust <- data.frame(uniquemodels, cutree(hc, k=500))
names(dfClust) <- c('modelname','cluster')
t <- table(dfClust$cluster)
t <- cbind(t,t/length(dfClust$cluster))
t <- t[order(t[,2], decreasing=TRUE),]
p <- data.frame(factorName=rownames(t), binCount=t[,1], percentFound=t[,2])
dfClust <- merge(x=dfClust, y=p, by.x = 'cluster', by.y='factorName', all.x=T)
dfClust <- dfClust[rev(order(dfClust$binCount)),]
names(dfClust) <- c('cluster','modelname')
head (dfClust[c('cluster','modelname')],50)
# build a convenient function to do all of the above
GroupFactorsTogether <- function(objData, variableName, clustersize=200, method='jw') {
# osa: Optimal string aligment, (restricted Damerau-Levenshtein distance).
# lv: Levenshtein distance (as in R's native adist).
# dl: Full Damerau-Levenshtein distance.
# hamming: Hamming distance (a and b must have same nr of characters).
# lcs: Longest common substring distance.
# qgram: q-gram distance.
# cosine: cosine distance between q-gram profiles
# jaccard: Jaccard distance between q-gram profiles
# jw: Jaro, or Jaro-Winker distance.
# soundex: Distance based on soundex encoding
# stringdistmatrix(a, b, method = c("osa", "lv", "dl", "hamming", "lcs",
# "qgram", "cosine", "jaccard", "jw", useBytes = FALSE,
# weight = c(d = 1, i = 1, s = 1, t = 1), maxDist = Inf, q = 1, p = 0,
# useNames = FALSE, ncores = 1, cluster = NULL)
# require(stringdist)
str <- unique(as.character(objData[,variableName]))
print(paste('Uniques:',length(str)))
d <- stringdistmatrix(str,str,method = c(method))
rownames(d) <- str
hc <- hclust(as.dist(d))
dfClust <- data.frame(str, cutree(hc, k=clustersize))
plot(table(dfClust$'cutree.hc..k...k.'))
most_populated_clusters <- dfClust[dfClust$'cutree.hc..k...k.' > 5,]
names(most_populated_clusters) <- c('entry','cluster')
# sort by most frequent
t <- table(most_populated_clusters$cluster)
t <- cbind(t,t/length(most_populated_clusters$cluster))
t <- t[order(t[,2], decreasing=TRUE),]
p <- data.frame(factorName=rownames(t), binCount=t[,1], percentFound=t[,2])
most_populated_clusters <- merge(x=most_populated_clusters, y=p, by.x = 'cluster', by.y='factorName', all.x=T)
most_populated_clusters <- most_populated_clusters[rev(order(most_populated_clusters$binCount)),]
names(most_populated_clusters) <- c('cluster','entry')
return (most_populated_clusters[c('cluster','entry')])
}
Manuel Amunategui - Follow me on Twitter: @amunategui

